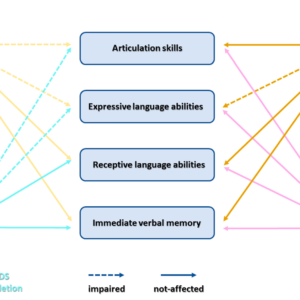
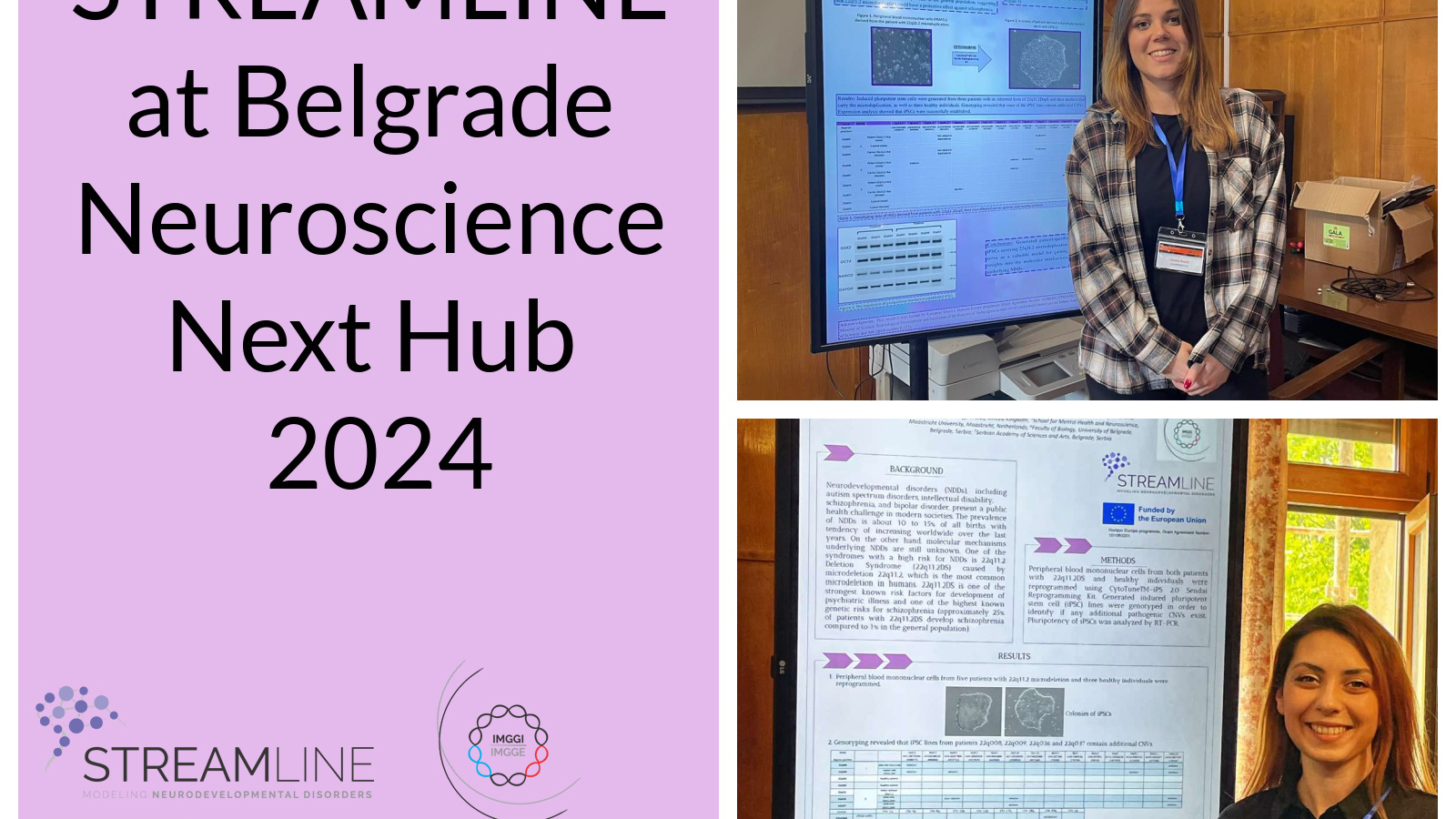
On Friday, May 31, 2024, the Institute for Biological Research “Siniša Stanković” – an Institute of national importance for the Republic of Serbia, University of Belgrade celebrated 77 years since the establishment. On this occasion, the Institute organized a traditional celebration. The first part of the celebration was the scientific part comprising of a series of lectures, which was followed by a festive part and awards ceremony. Dr Danijela Drakulić had a great honour to present STREAMLINE project as an esteemed guest during scientific segment of this program.